Project participants
Dr. Nolwenn Briand, Sarah Hazel Pickering (PhD student; former), Dr. Natalia M. Galigniana, Dr. Julia Madsen Østerbye, Dr. Mohamed Abdelhalim (bioinformatics)
Ongoing research
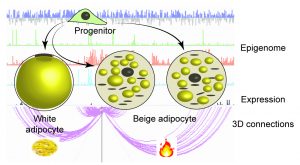
- Genome conformation changes and transcription factor dynamics during white and beige adipogenesis, using human cell culture model
- Interplay between the nuclear lamina, LADs and TADs in the differentiation of adipose stem cells
- Regulation of gene expression at the nuclear lamina
Recent findings
- Cytoskeletal and nucleolar rearrangements during adipogenesis (Potolitsyna, Hazell Pickering 2024) Commun Biol
- Enhancer connectivity and gene activity at the nuclear lamina during adipocyte differentiation Madsen-Østerbye 2022 Genome Biol (Madsen-Østerbye 2023 Genes)
- 'Pockets' of euchromatin and active genes in LADs (Madsen-Østerbye 2022 Genome Biol)
- A polymer model of chromatin to infer chromatin behavior at the nuclear lamina (Brunet 2021 Nucleus)
- TAD cliques shape the 4D genome during adipose differentiation (Paulsen 2019 Nature Genet)
- Chrom3D: a platform for 3D genome modeling from Hi-C and LAD data (Paulsen 2017 Genome Biol; Paulsen 2018 Nature Protoc)
- Chrom3D on github: https://github.com/Chrom3D/Chrom3D
- ChIP protocol for nuclear lamins (Oldenburg 2016 Method Mol Biol)
- Constitutive and facultative LADs during adipogenic differentiation (Rønningen 2015 Genome Res)
- Enriched Domain Detector (EDD) to identify LADs and other broad genomic domains (Lund 2014 Nucl Acids Res)
- EDD on github: https://github.com/CollasLab/edd
- Dynamic interactions of lamin A/C with promoters during adipogenic differentiation (Lund 2013 Genome Res)